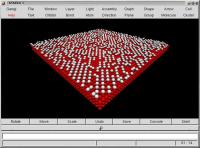
Maura E. Monville, Zhenyu Zhang and George H.Gilmer,
Monte Carlo simulation of clustering effects on surfaces,
Oak Ridge National Laboratory and Lawrence Livermore National
Laboratory, USA. This image shows 1030 adatoms over 7500 substrate
atoms, after 20,000 simulation cycles. Size: 92,832 bytes.
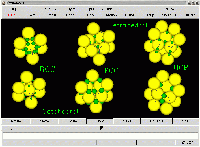
BCC, FCC and HCP structural cells (yellow atoms), showing all the tetrahedral
and octahedral interstices available, as described by the largest atoms that
can occupy those sites without causing distortion (green atoms). Atoms and Cells
are all represented on the same scale, structural (yellow) atoms have all
a radius equal to 1.0, and cell parameters were chosen so structural atoms
just touch the first neighbours. A few structural atoms were removed to
improve visibility. Size: 46,189 bytes.
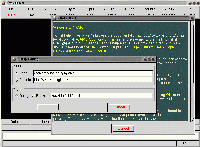
The Help->Config dialog, to configure documentation source
(local or remote, HTTP or FTP) and carrying agent (Gamgi or a Browser),
over the Help->Start dialog, saluting newbies (using the same
green and yellow default colors as Emacs on Red Hat 9.0).
Help documentation is taken from XML files, except Help->Start
which is directly built-in (so it is always available, even if
documentation paths are misconfigured). Size: 49,379 bytes.

Maura E. Monville, Zhenyu Zhang and George H.Gilmer,
Oak Ridge National Laboratory and Lawrence Livermore National Laboratory, USA.
Atomic deposition over a substrate lattice, by Monte Carlo simulation,
as a result of the combined effect of Laser Steering and Temperature Gradient.
Both effects stem from the interference pattern (standing wave) formed
by two counter-propagating waves (the two laser beams E-fields respectively).
The goal is to prove that by proper tuning of the Laser
experiment-controllable parameters the two effects can be made work
together in a constructive way so enhancing the resolution of nano-scale
atomic patterns in cases of deposition of metal on metal. Size: 161,387 bytes.
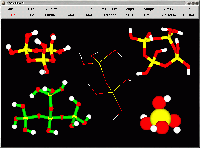
Some of the silica clusters acting as building units in sol-gel processes,
zeolite catalysis and in the chemistry of silica in solution, shown with
different styles. The dimer Si2O(OH)6, represented in wired mode, was built
on a different, transparent, layer, so it is not affected by the diffuse
light used to illuminate the other clusters. The monomer Si(OH)4, at the
bottom right corner, was represented using the tabulated covalent radius
for Si, O, H. The bottom menus were switched off, to emphasize the image.
Size: 37,945 bytes.
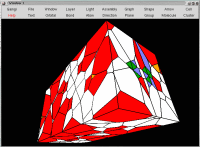
Voronoi tesselation of the
DNA double-helix (698 atoms), limited by
six outer boundary planes. Silicons are yellow, Oxygens are red, Nitrogens
are blue, Carbons are green, Phosporous are orange and Hydrogens are white.
Size: 30,099 bytes.
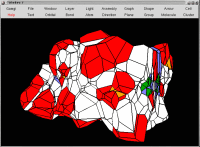
Voronoi tesselation of the
DNA double-helix (698 atoms), using
periodic boundary conditions, with the extended convention (the minimum
convention, usually applied in Molecular Dynamics, cannot be used in
Voronoi tesselation). Silicons are yellow, Oxygens are red, Nitrogens
are blue, Carbons are green, Phosporous are orange and Hydrogens are white.
Size: 34,100 bytes.
 Maura E. Monville, Zhenyu Zhang and George H.Gilmer,
Monte Carlo simulation of clustering effects on surfaces,
Oak Ridge National Laboratory and Lawrence Livermore National
Laboratory, USA. This image shows 1030 adatoms over 7500 substrate
atoms, after 20,000 simulation cycles. Size: 92,832 bytes.
Maura E. Monville, Zhenyu Zhang and George H.Gilmer,
Monte Carlo simulation of clustering effects on surfaces,
Oak Ridge National Laboratory and Lawrence Livermore National
Laboratory, USA. This image shows 1030 adatoms over 7500 substrate
atoms, after 20,000 simulation cycles. Size: 92,832 bytes.
 BCC, FCC and HCP structural cells (yellow atoms), showing all the tetrahedral
and octahedral interstices available, as described by the largest atoms that
can occupy those sites without causing distortion (green atoms). Atoms and Cells
are all represented on the same scale, structural (yellow) atoms have all
a radius equal to 1.0, and cell parameters were chosen so structural atoms
just touch the first neighbours. A few structural atoms were removed to
improve visibility. Size: 46,189 bytes.
BCC, FCC and HCP structural cells (yellow atoms), showing all the tetrahedral
and octahedral interstices available, as described by the largest atoms that
can occupy those sites without causing distortion (green atoms). Atoms and Cells
are all represented on the same scale, structural (yellow) atoms have all
a radius equal to 1.0, and cell parameters were chosen so structural atoms
just touch the first neighbours. A few structural atoms were removed to
improve visibility. Size: 46,189 bytes.
 The Help->Config dialog, to configure documentation source
(local or remote, HTTP or FTP) and carrying agent (Gamgi or a Browser),
over the Help->Start dialog, saluting newbies (using the same
green and yellow default colors as Emacs on Red Hat 9.0).
Help documentation is taken from XML files, except Help->Start
which is directly built-in (so it is always available, even if
documentation paths are misconfigured). Size: 49,379 bytes.
The Help->Config dialog, to configure documentation source
(local or remote, HTTP or FTP) and carrying agent (Gamgi or a Browser),
over the Help->Start dialog, saluting newbies (using the same
green and yellow default colors as Emacs on Red Hat 9.0).
Help documentation is taken from XML files, except Help->Start
which is directly built-in (so it is always available, even if
documentation paths are misconfigured). Size: 49,379 bytes.
 Maura E. Monville, Zhenyu Zhang and George H.Gilmer,
Oak Ridge National Laboratory and Lawrence Livermore National Laboratory, USA.
Atomic deposition over a substrate lattice, by Monte Carlo simulation,
as a result of the combined effect of Laser Steering and Temperature Gradient.
Both effects stem from the interference pattern (standing wave) formed
by two counter-propagating waves (the two laser beams E-fields respectively).
The goal is to prove that by proper tuning of the Laser
experiment-controllable parameters the two effects can be made work
together in a constructive way so enhancing the resolution of nano-scale
atomic patterns in cases of deposition of metal on metal. Size: 161,387 bytes.
Maura E. Monville, Zhenyu Zhang and George H.Gilmer,
Oak Ridge National Laboratory and Lawrence Livermore National Laboratory, USA.
Atomic deposition over a substrate lattice, by Monte Carlo simulation,
as a result of the combined effect of Laser Steering and Temperature Gradient.
Both effects stem from the interference pattern (standing wave) formed
by two counter-propagating waves (the two laser beams E-fields respectively).
The goal is to prove that by proper tuning of the Laser
experiment-controllable parameters the two effects can be made work
together in a constructive way so enhancing the resolution of nano-scale
atomic patterns in cases of deposition of metal on metal. Size: 161,387 bytes.
 Some of the silica clusters acting as building units in sol-gel processes,
zeolite catalysis and in the chemistry of silica in solution, shown with
different styles. The dimer Si2O(OH)6, represented in wired mode, was built
on a different, transparent, layer, so it is not affected by the diffuse
light used to illuminate the other clusters. The monomer Si(OH)4, at the
bottom right corner, was represented using the tabulated covalent radius
for Si, O, H. The bottom menus were switched off, to emphasize the image.
Size: 37,945 bytes.
Some of the silica clusters acting as building units in sol-gel processes,
zeolite catalysis and in the chemistry of silica in solution, shown with
different styles. The dimer Si2O(OH)6, represented in wired mode, was built
on a different, transparent, layer, so it is not affected by the diffuse
light used to illuminate the other clusters. The monomer Si(OH)4, at the
bottom right corner, was represented using the tabulated covalent radius
for Si, O, H. The bottom menus were switched off, to emphasize the image.
Size: 37,945 bytes.
 Voronoi tesselation of the
DNA double-helix (698 atoms), limited by
six outer boundary planes. Silicons are yellow, Oxygens are red, Nitrogens
are blue, Carbons are green, Phosporous are orange and Hydrogens are white.
Size: 30,099 bytes.
Voronoi tesselation of the
DNA double-helix (698 atoms), limited by
six outer boundary planes. Silicons are yellow, Oxygens are red, Nitrogens
are blue, Carbons are green, Phosporous are orange and Hydrogens are white.
Size: 30,099 bytes.
 Voronoi tesselation of the
DNA double-helix (698 atoms), using
periodic boundary conditions, with the extended convention (the minimum
convention, usually applied in Molecular Dynamics, cannot be used in
Voronoi tesselation). Silicons are yellow, Oxygens are red, Nitrogens
are blue, Carbons are green, Phosporous are orange and Hydrogens are white.
Size: 34,100 bytes.
Voronoi tesselation of the
DNA double-helix (698 atoms), using
periodic boundary conditions, with the extended convention (the minimum
convention, usually applied in Molecular Dynamics, cannot be used in
Voronoi tesselation). Silicons are yellow, Oxygens are red, Nitrogens
are blue, Carbons are green, Phosporous are orange and Hydrogens are white.
Size: 34,100 bytes.